Spectral Permutation Demo
Importing Packages
import numpy as np
import pandas as pd
from mheatmap import (
amc_postprocess,
mosaic_heatmap
)
from mheatmap.graph import spectral_permute
from mheatmap.constants import set_test_mode
set_test_mode(True)
import matplotlib.pyplot as plt
import scipy
Load Data
-
Load the ground truth labels
Salinas_gt.mat: Ground truth labels for Salinas dataset
-
Load the predicted labels from
spectral clustering
# Load the data
y_true = scipy.io.loadmat("data/Salinas_gt.mat")["salinas_gt"].reshape(-1)
# Load predicted labels from spectral clustering
y_pred = np.array(
pd.read_csv(
"data/Salinas_spectralclustering.csv",
header=None,
low_memory=False,
)
.values[1:]
.flatten()
)
print(f"y_true shape: {y_true.shape}")
print(f"y_pred shape: {len(y_pred)}")
y_true shape: (111104,)
y_pred shape: 111104
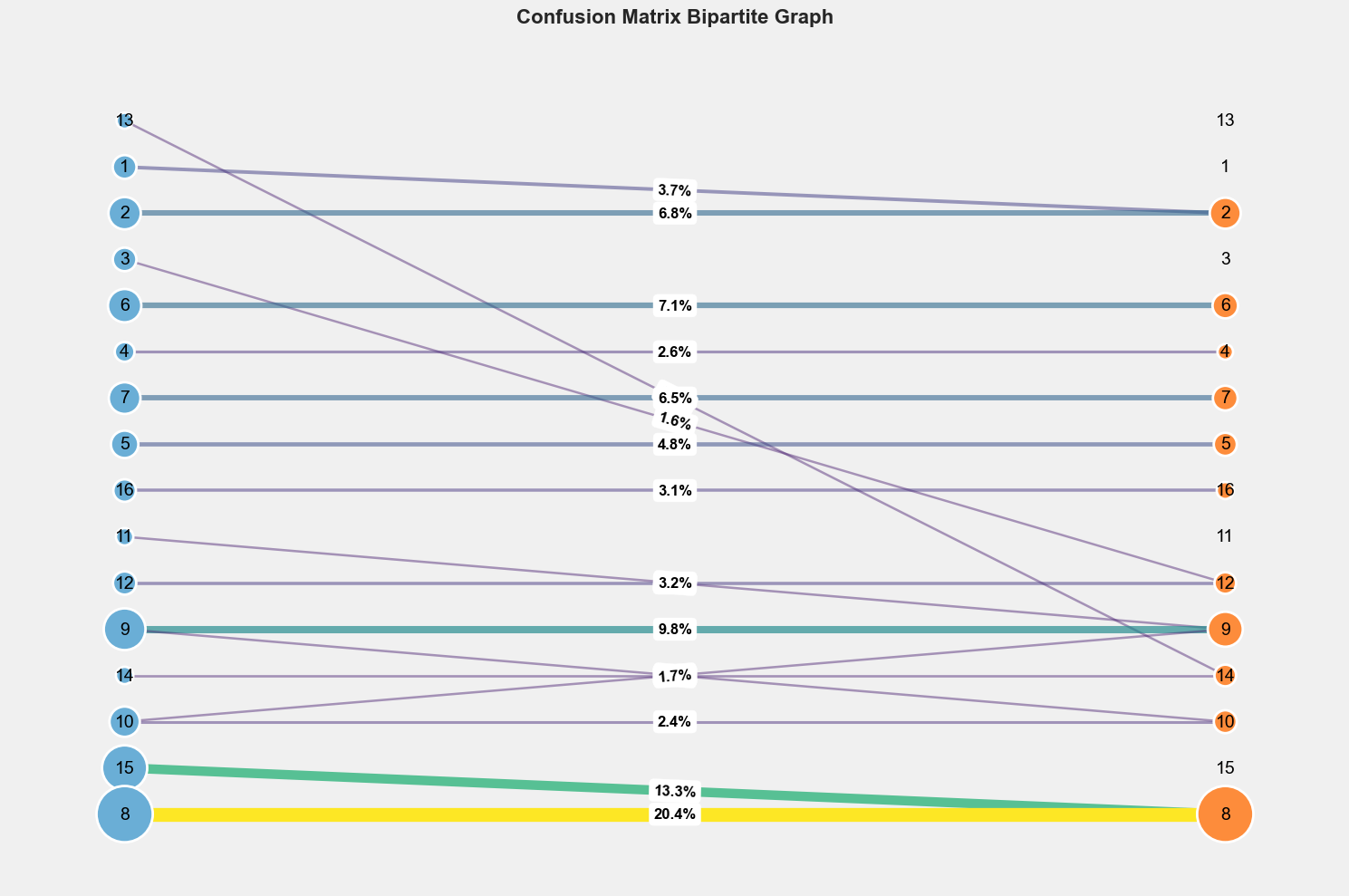
AMC Post-processing
- Alignment with
Hungarianalgorithm - Masking the zeros (unlabeled pixels) with mask_zeros_from_gt
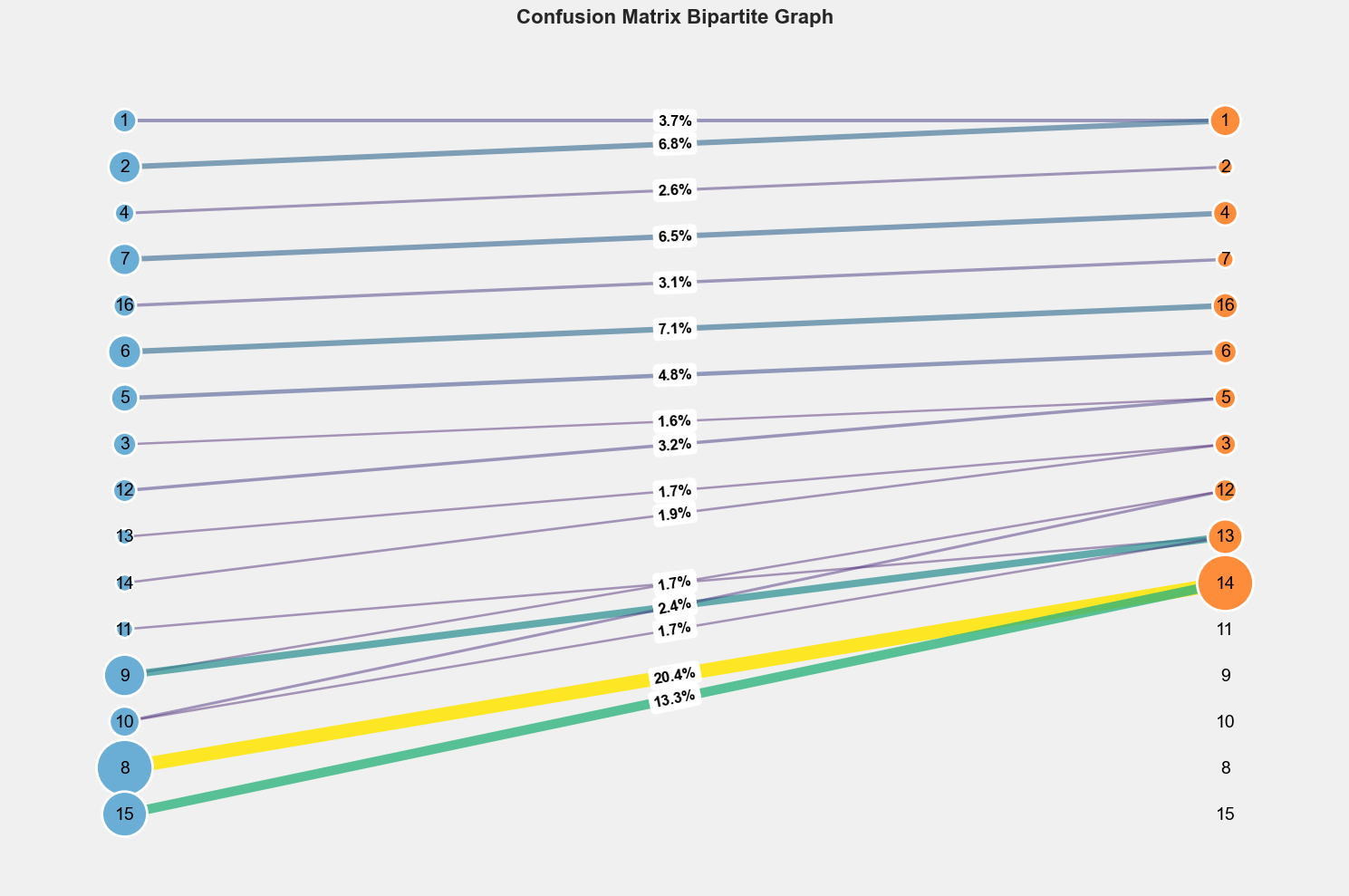
- Computing the confusion matrix
See AMC Post-processing for more details.
Spectral Permutation
- Treat the confusion matrix as an adjacency matrix
\[
A = C
\]
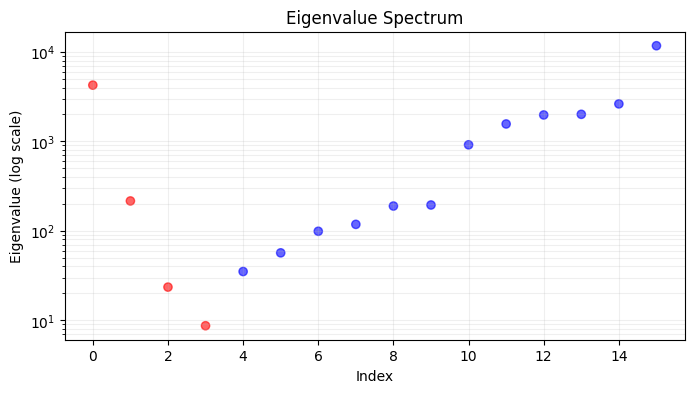
- Compute the graph Laplacian
\[
L = D - A
\]
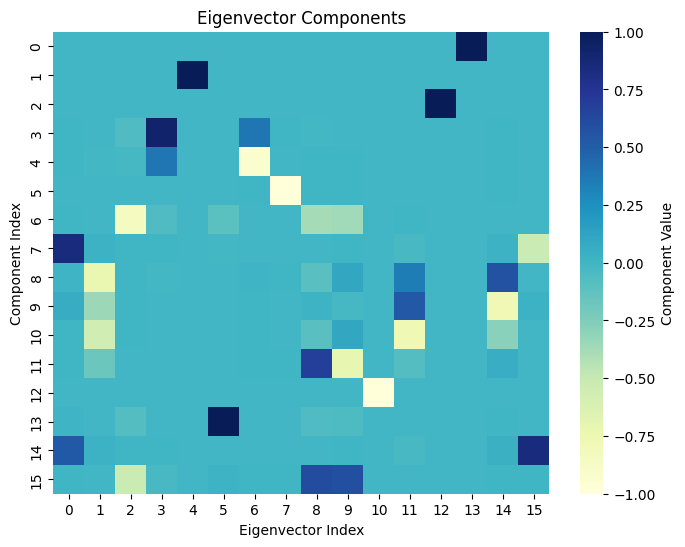
- Compute the eigenvectors and eigenvalues
\[
L = U \Sigma U^T
\]
- Sort the eigenvectors based on the eigenvalues
\[
U = [u_1, u_2, \ldots, u_n]
\]
- Fiedler vector is the second smallest eigenvector
\[
f = u_2
\]
- Sort the labels based on the Fiedler vector
\[
\pi\tilde{y} = \tilde{y}[f]
\]
- Reorder the confusion matrix based on the sorted labels
\[
\pi C = C[f, f]
\]
Visualize the Results
# Visualize original vs spectrally reordered matrices
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 6))
mosaic_heatmap(conf_mat, ax=ax1, xticklabels=labels, yticklabels=labels, cmap="YlGnBu")
ax1.set_title("Original", fontsize=18, color='#4A4A4A') # Medium gray
ax1.xaxis.set_ticks_position('top')
ax1.tick_params(colors='#4A4A4A')
mosaic_heatmap(
reordered_mat,
ax=ax2,
xticklabels=reordered_labels,
yticklabels=reordered_labels,
cmap="YlGnBu",
)
ax2.set_title("Spectral Reordered", fontsize=18, color='#4A4A4A') # Medium gray
ax2.xaxis.set_ticks_position('top')
ax2.tick_params(colors='#4A4A4A')
plt.tight_layout()
plt.show()
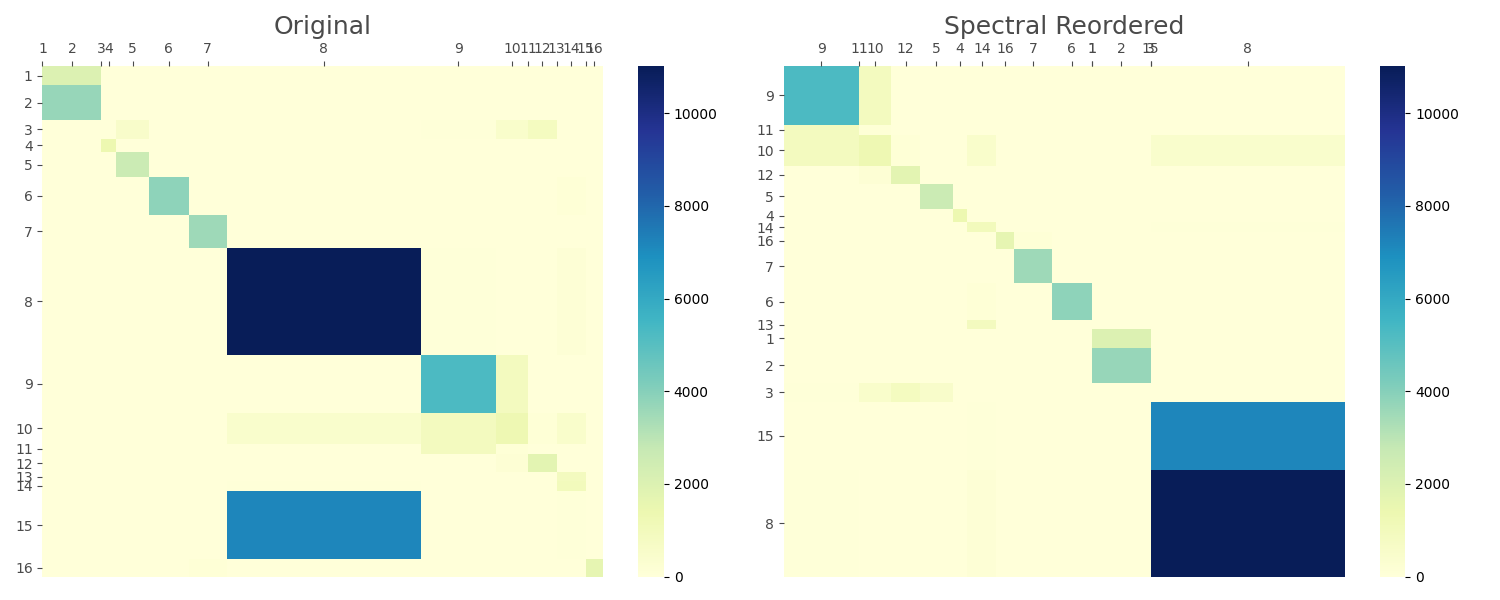
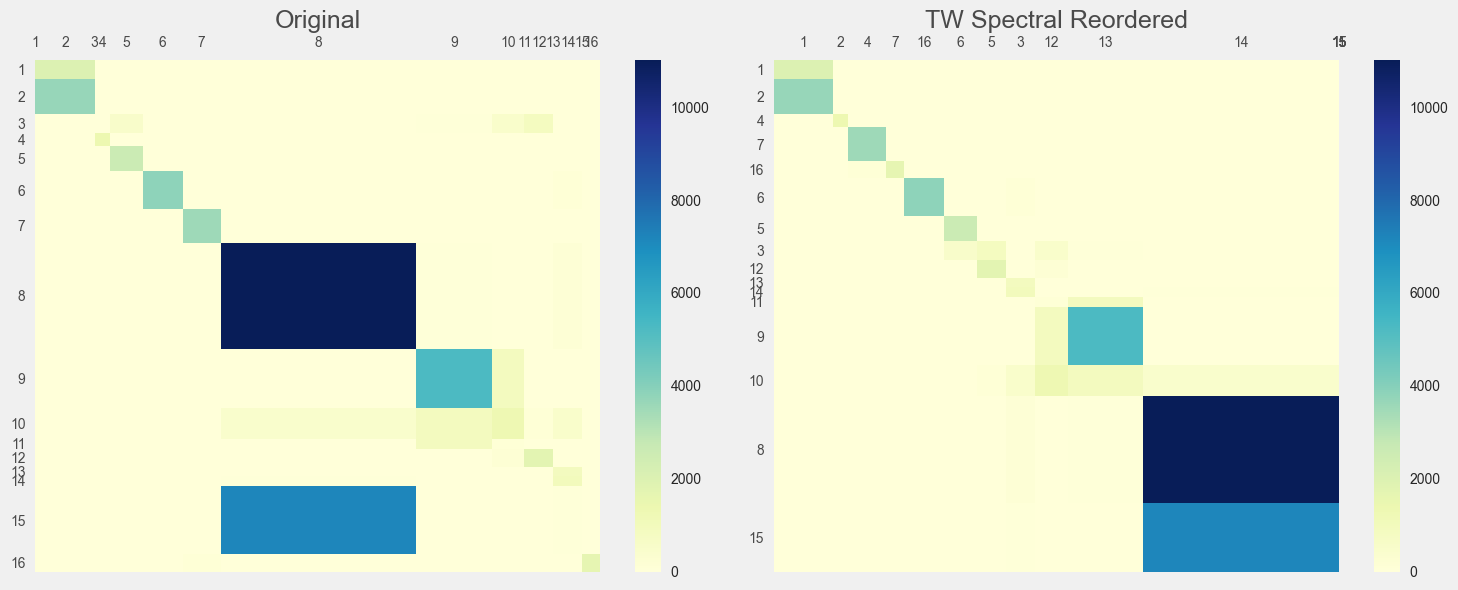
Two-walk Spectral Permutation
# Two-walk Laplacian
reordered_mat, reordered_labels = spectral_permute(conf_mat, labels, mode='tw')
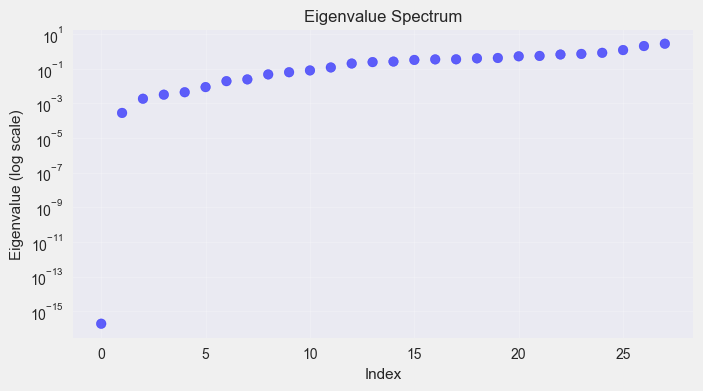
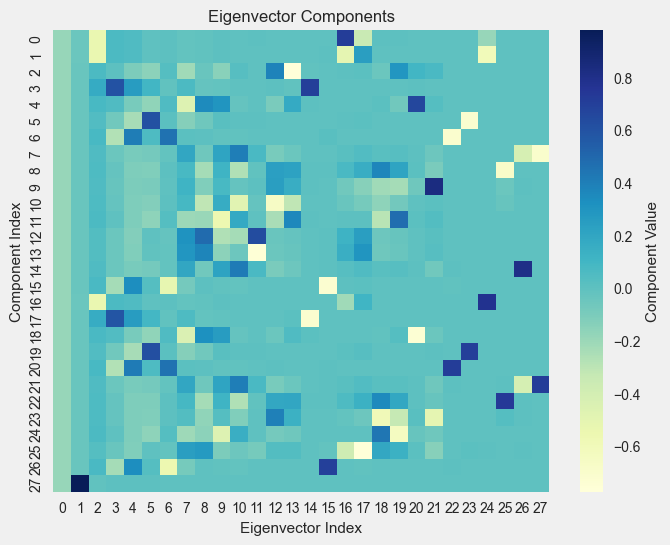
Visualize the TW Results
# Visualize original vs spectrally reordered matrices
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 6))
mosaic_heatmap(conf_mat, ax=ax1, xticklabels=labels, yticklabels=labels, cmap="YlGnBu")
ax1.set_title("Original", fontsize=18, color='#4A4A4A') # Medium gray
ax1.xaxis.set_ticks_position('top')
ax1.tick_params(colors='#4A4A4A')
mosaic_heatmap(
reordered_mat,
ax=ax2,
xticklabels=reordered_labels,
yticklabels=reordered_labels,
cmap="YlGnBu",
)
ax2.set_title("TW Spectral Reordered", fontsize=18, color='#4A4A4A') # Medium gray
ax2.xaxis.set_ticks_position('top')
ax2.tick_params(colors='#4A4A4A')
plt.tight_layout()
plt.show()